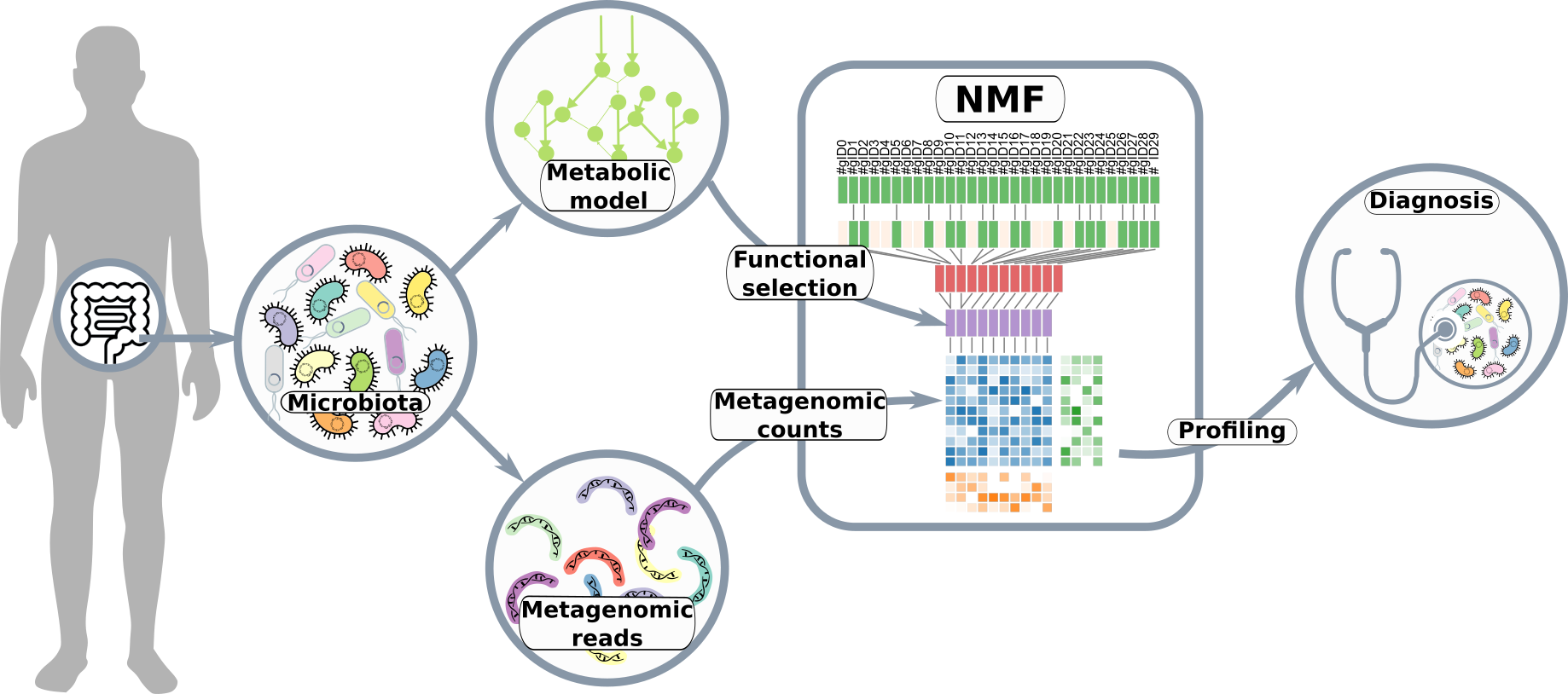
Four functional profiles for fibre and mucin metabolism in the human gut microbiome
Conference
65th ISI World Statistics Congress
Format: IPS Abstract - WSC 2025
Keywords: metagenomics, microbiota
Session: IPS 927 - Statistical Tools for Microbiome-Based Biomarker Identification and Disease Prediction
Thursday 9 October 2 p.m. - 3:40 p.m. (Europe/Amsterdam)
Abstract
With the emergence of metagenomic data, multiple links between the gut microbiome and the host health have been shown. Deciphering these complex interactions require evolved analysis methods focusing on the microbial ecosystem functions. Despite the fact that host or diet-derived fibres are the most abundant nutrients available in the gut, the presence of distinct functional traits regarding fibre and mucin hydrolysis, fermentation and hydrogenotrophic processes has never been investigated.
After manually selecting 91 KEGG orthologies and 33 glycoside hydrolases further aggregated in 101 functional descriptors representative of fibre and mucin degradation pathways in the gut microbiome, we used nonnegative matrix factorization (NMF) to mine metagenomic datasets. The NMF method has been supplemented by functional constrained and L1-L2 constraints and hyperparameters were thoroughly selected with three different criteria. Four distinct metabolic profiles were identified on a training set of 1153 samples, and validated on a large database of 2571 unseen samples from 5 external metagenomic cohorts.
The functional exploration of these profiles indicates that Profiles 1 and 2 are the main contributors to the fibre-degradation-related metagenome: they present contrasted involvement in fibre degradation and sugar metabolism and are differentially linked to dysbiosis, metabolic disease and inflammation. Profile 1 takes over Profile 2 in healthy samples, and unbalance of these profiles characterize dysbiotic samples. Furthermore, high fibre diet favours a healthy balance between profiles 1 and profile 2. Profile 3 takes over profile 2 during Crohn’s disease, inducing functional reorientations towards unusual metabolism. Profile 4 gathers under-represented functions, like methanogenesis. Two taxonomic makes up of the profiles were investigated using non-negative regression, using either the covariation of 203 prevalent genomes or metagenomic species, both providing consistent results in line with their functional characteristics.
Conclusions: integrating anaerobic microbiology knowledge with statistical learning can narrow down the metagenomic analysis to investigate functional profiles. Applying this approach to fibre degradation in the gut ended with 4 distinct functional profiles that can be easily monitored as markers of diet, dysbiosis, inflammation and disease.
Figures/Tables
outline